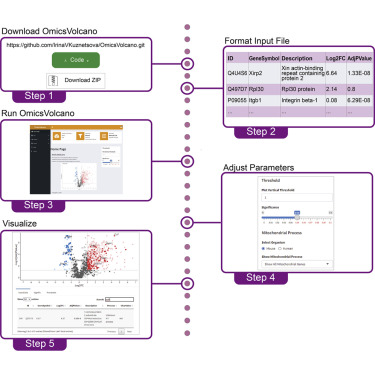
Advances in omics technologies have generated exponentially larger volumes of biological data; however, their analyses and interpretation are limited to computationally proficient scientists. We created OmicsVolcano, an interactive open-source software tool to enable visualization and exploration of high-throughput biological data, while highlighting features of interest using a volcano plot interface. In contrast to existing tools, our software and user-interface design allow it to be used without requiring any programming skills to generate high-quality and presentation-ready images.
The software tool is freely available on GitHub – [1]. Our article is open access, and can be download from [2].
- Collaborators: Harry Perkins Institute of Medical Research, University of Western Australia (UWA), AUSTRALIA; Umea University (UMU), Umea, Sweden; and Edith Cowan University (ECU), AUSTRALIA
- Authors: Irina Kuznetsova, Artur Lugmayr, Stefan Siira, Oliver Rackham, Aleksandra Filipovska
- Publications: [2]
- GitHub: https://github.com/IrinaVKuznetsova/CirGO.git
[Bibtex]
@WWW{Lugmayr:2019:CirGo:GitHub,
author = {Kuznetsova, Irina and Lugmayr, Artur and Siira, Stefan J. and Rackham, Oliver and Filipovska, Aleksandra},
title = {CirGO: an alternative circular way of visualising gene ontology terms (GitHub): https://github.com/IrinaVKuznetsova/CirGO},
url = {https://github.com/IrinaVKuznetsova/CirGO},
}![[pdf]](http://artur-lugmayr.com/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@Article{Lugmayr:2019:CirGo,
author = {Kuznetsova, Irina and Lugmayr, Artur and Siira, Stefan J. and Rackham, Oliver and Filipovska, Aleksandra},
title = {CirGO: an alternative circular way of visualising gene ontology terms},
doi = {10.1186/s12859-019-2671-2},
issn = {1471-2105},
number = {1},
pages = {84},
url = {https://doi.org/10.1186/s12859-019-2671-2},
volume = {20},
abstract = {Prioritisation of gene ontology terms from differential gene expression
analyses in a two-dimensional format remains a challenge with exponentially
growing data volumes. Typically, gene ontology terms are represented
as tree-maps that enclose all data into defined space. However, large
datasets make this type of visualisation appear cluttered and busy,
and often not informative as some labels are omitted due space limits,
especially when published in two-dimensional (2D) figures.},
day = {18},
file = {:Lugmayr_2019_CirGo - CirGO_ an Alternative Circular Way of Visualising Gene Ontology Terms.pdf:PDF},
journal = {BMC Bioinformatics},
month = {Feb},
year = {2019},
}